
Support Vector Machines | Part II
Non-linear Kernels
Some common non-linear kernels used for SVM include polynomial and radial basis function (RBF). You can find additional information about what kernels can be used on the SciKit Learn website
https://scikit-learn.org/stable/auto_examples/svm/plot_svm_kernels.html
Start by normalizing the data. To normalize data, it is best to use a pipeline because this first scales the training data and then applies the same scale to the test data without leading any training data.
# Import required functions
from sklearn.datasets import make_classification
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import train_test_split
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import StandardScaler
# Split dataset into training set and test set
X_train, X_test, y_train, y_test = train_test_split(wine.data, wine.target, test_size=0.3,random_state=109) # 70% training and 30% test
# Make pipeline for normalizing data
pipe = make_pipeline(StandardScaler(), LogisticRegression())
pipe.fit(X_train, y_train) # apply scaling on training data
pipe.score(X_test, y_test) # apply scaling on testing data, without leaking training data.
0.9259259259259259
Radial Basis Function (RBF) Kernel
SVM has a parameter called C which can be adjusted to allow for a certain level of mis-classification to have a more optimized location of the decision boundary.
When using the RBF kernel, there is another parameter, gamma, which can be adjusted to change how linear the boundary is.
From https://scikit-learn.org/stable/auto_examples/svm/plot_rbf_parameters.html:
"The gamma parameter defines how far the influence of a single training example reaches, with low values meaning ‘far’ and high values meaning ‘close’.
To find the best gamma and C values for RBF, it is a good idea to do an initial search. For this initial search, it is possible to use a logarithmic grid with basis 10.
from sklearn.svm import SVC
from sklearn.model_selection import StratifiedShuffleSplit
from sklearn.model_selection import GridSearchCV
C_range = np.logspace(-2, 10, 13)
gamma_range = np.logspace(-9, 3, 13)
param_grid = dict(gamma=gamma_range, C=C_range)
cv = StratifiedShuffleSplit(n_splits=5, test_size=0.2, random_state=42)
grid = GridSearchCV(SVC(), param_grid=param_grid, cv=cv)
grid.fit(X_train, y_train)
print("The best parameters are", grid.best_params_, "with a score of", grid.best_score_)
The best parameters are {'C': 100000.0, 'gamma': 1e-07} with a score of 0.944
Train the model based on the gamma and C values that were determined above.
#Import svm model
from sklearn import svm
#Create a svm Classifier
clf = svm.SVC(kernel="rbf", gamma=1e-07, C=100000.0) # RBF Kernel
#Train the model using the training sets
clf.fit(X_train, y_train)
#Predict the response for test dataset
y_pred = clf.predict(X_test)
#Import scikit-learn metrics module for accuracy calculation
from sklearn import metrics
# Model Accuracy: how often is the classifier correct?
print("Accuracy:",metrics.accuracy_score(y_test, y_pred))
# Which classes are commonly misclassified?
print('Confusion Matrix')
print(metrics.confusion_matrix(y_test, y_pred, labels=None))
Accuracy: 0.8703703703703703
Confusion Matrix
[[19 2 0]
[ 4 15 0]
[ 0 1 13]]
We can again visualize the decision boundary on the first 2 features of the dataset. It will take a few more seconds to run as it processes the data.
from sklearn.svm import SVC
import numpy as np
import matplotlib.pyplot as plt
from sklearn import svm, datasets
# Select 2 features / variable for the 2D plot that we are going to create.
X = wine.data[:, :2] # we only take the first two features.
y = wine.target
def make_meshgrid(x, y, h=.02):
x_min, x_max = x.min() - 1, x.max() + 1
y_min, y_max = y.min() - 1, y.max() + 1
xx, yy = np.meshgrid(np.arange(x_min, x_max, h), np.arange(y_min, y_max, h))
return xx, yy
def plot_contours(ax, clf, xx, yy, **params):
Z = clf.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
out = ax.contourf(xx, yy, Z, **params)
return out
model = svm.SVC(kernel='rbf', gamma=1e-07, C=100000.0)
clf = model.fit(X, y)
fig, ax = plt.subplots()
# create a mesh to plot in
h = .02 # step size in the mesh
x_min, x_max = X[:, 0].min() - 1, X[:, 0].max() + 1
y_min, y_max = X[:, 1].min() - 1, X[:, 1].max() + 1
xx, yy = np.meshgrid(np.arange(x_min, x_max, h),
np.arange(y_min, y_max, h))
Z = clf.predict(np.c_[xx.ravel(), yy.ravel()])
# Put the result into a color plot
Z = Z.reshape(xx.shape)
plt.contourf(xx, yy, Z, cmap=plt.cm.coolwarm, alpha=0.8)
# Plot also the training points
plt.scatter(X[:, 0], X[:, 1], c=y, cmap=plt.cm.coolwarm, edgecolors='k')
plt.xlabel('Alcohol')
plt.ylabel('Malic Acid')
plt.xlim(xx.min(), xx.max())
plt.ylim(yy.min(), yy.max())
plt.xticks(())
plt.yticks(())
plt.title("Decision Surface of SVM using RBF Kernel")
plt.show()
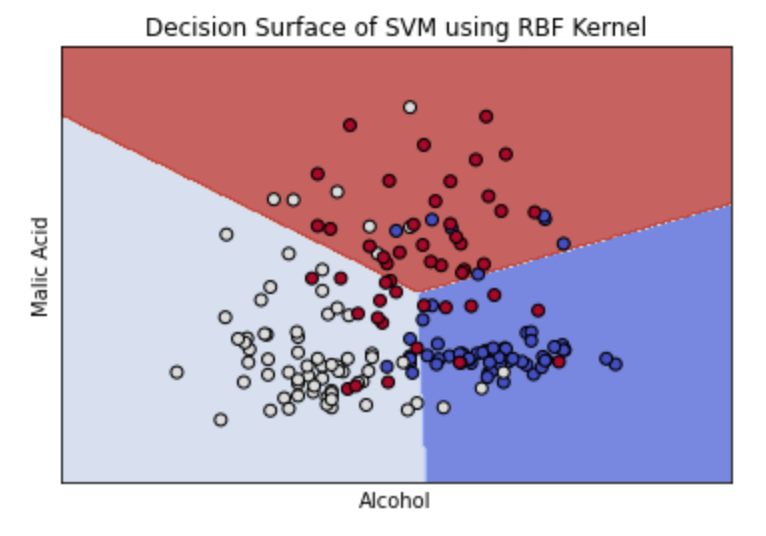
The graph above shows how the optimal parameters using the RBF kernel created an almost linear separation between the data. If we increase the gamma value, the separation boundary creates curves around specific groups of points. See this in the example below. For the sample dataset, this is less accurate. Additionally, higher gamma values can lead to overfitting.
from sklearn.svm import SVC import numpy as np import matplotlib.pyplot as plt from sklearn import svm, datasets # Select 2 features / variable for the 2D plot that we are going to create. X = wine.data[:, :2] # we only take the first two features. y = wine.target def make_meshgrid(x, y, h=.02): x_min, x_max = x.min() - 1, x.max() + 1 y_min, y_max = y.min() - 1, y.max() + 1 xx, yy = np.meshgrid(np.arange(x_min, x_max, h), np.arange(y_min, y_max, h)) return xx, yy def plot_contours(ax, clf, xx, yy, **params): Z = clf.predict(np.c_[xx.ravel(), yy.ravel()]) Z = Z.reshape(xx.shape) out = ax.contourf(xx, yy, Z, **params) return out model = svm.SVC(kernel='rbf', gamma=5, C=100000.0) clf = model.fit(X, y) fig, ax = plt.subplots()
# create a mesh to plot in h = .02 # step size in the mesh x_min, x_max = X[:, 0].min() - 1, X[:, 0].max() + 1 y_min, y_max = X[:, 1].min() - 1, X[:, 1].max() + 1 xx, yy = np.meshgrid(np.arange(x_min, x_max, h), np.arange(y_min, y_max, h)) Z = clf.predict(np.c_[xx.ravel(), yy.ravel()]) # Put the result into a color plot Z = Z.reshape(xx.shape) plt.contourf(xx, yy, Z, cmap=plt.cm.coolwarm, alpha=0.8) # Plot also the training points plt.scatter(X[:, 0], X[:, 1], c=y, cmap=plt.cm.coolwarm, edgecolors='k') plt.xlabel('Alcohol') plt.ylabel('Malic Acid') plt.xlim(xx.min(), xx.max()) plt.ylim(yy.min(), yy.max()) plt.xticks(()) plt.yticks(()) plt.title("Decision Surface of SVM using RBF Kernel") plt.show()
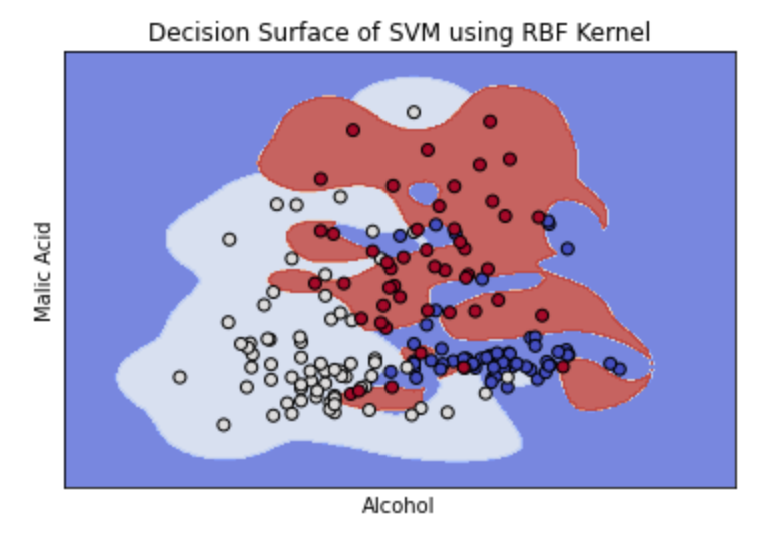
TASK: Plot a validation curve for the gamma value through SciKit Learn to see how the value of gamma impacts the accuracy of the model.
Polynomial kernel
The polynomial kernel has three parameters: degree, gamma, and C. The most common degree used is either 2 or 3 because larger degrees are more likely to overfit. Gamma can be set to "auto" which means it uses 1 / n_features. C is the same as the C used for the RBF kernel.
#Import svm model
from sklearn import svm
#Create a svm Classifier
clf = svm.SVC(kernel="poly", degree=2, gamma="auto", C=100000.0) # Polynomial Kernel
#Train the model using the training sets
clf.fit(X_train, y_train)
#Predict the response for test dataset
y_pred = clf.predict(X_test)
#Import scikit-learn metrics module for accuracy calculation
from sklearn import metrics
# Model Accuracy: how often is the classifier correct?
print("Accuracy:",metrics.accuracy_score(y_test, y_pred))
# Which classes are commonly misclassified?
print('Confusion Matrix')
print(metrics.confusion_matrix(y_test, y_pred, labels=None))
Accuracy: 0.9259259259259259
Confusion Matrix
[[21 0 0]
[ 2 16 1]
[ 1 0 13]]
from sklearn.svm import SVC
import numpy as np
import matplotlib.pyplot as plt
from sklearn import svm, datasets
# Select 2 features / variable for the 2D plot that we are going to create.
X = wine.data[:, :2] # we only take the first two features.
y = wine.target
def make_meshgrid(x, y, h=.02):
x_min, x_max = x.min() - 1, x.max() + 1
y_min, y_max = y.min() - 1, y.max() + 1
xx, yy = np.meshgrid(np.arange(x_min, x_max, h), np.arange(y_min, y_max, h))
return xx, yy
def plot_contours(ax, clf, xx, yy, **params):
Z = clf.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
out = ax.contourf(xx, yy, Z, **params)
return out
model = svm.SVC(kernel='rbf')
clf = model.fit(X, y)
fig, ax = plt.subplots()
# Set-up grid for plotting.
X0, X1 = X[:, 0], X[:, 1]
xx, yy = make_meshgrid(X0, X1)
plot_contours(ax, clf, xx, yy, cmap=plt.cm.coolwarm, alpha=0.8)
ax.scatter(X0, X1, c=y, cmap=plt.cm.coolwarm, s=20, edgecolors='k')
ax.set_xlabel('Alcohol')
ax.set_ylabel('Malic Acid')
ax.set_title('Decision Surface of SVM with Polynomial Kernel')
plt.show()
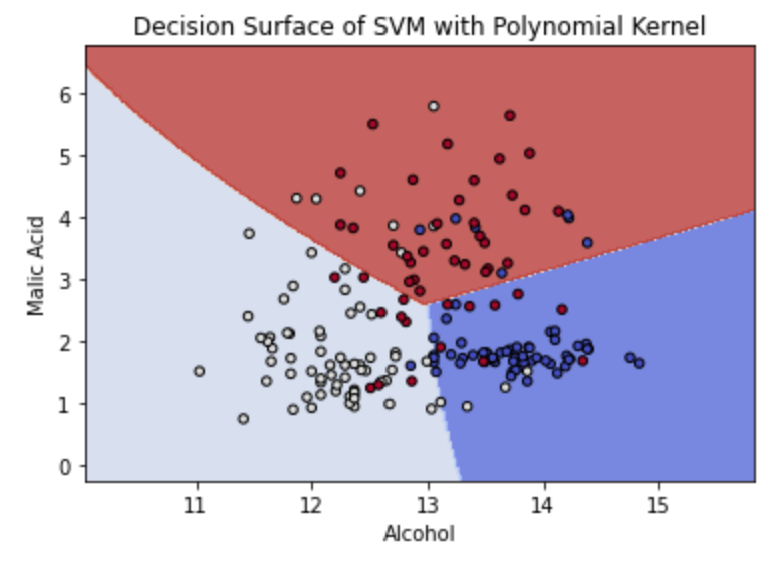
TASK: Plot a validation curve for the degree value through SciKit Learn to see how the number of degrees impacts the accuracy of the model.